Location:Home >> Detail
TOTAL VIEWS
J Psychiatry Brain Sci. 2017; 2(1): 2; https://doi.org/10.20900/jpbs.20170002
1 Department of Neurology, The First People's Hospital Affiliated to Soochow University, Suzhou, Jiangsu Province, 215006, China
2 Department of Neurology, Jiangyin People's hospital. Jiangyin, Jiangsu Province, 214400, China
3 Department of Statistics, Rice University, 6100 Main St. Duncan Hall 2124, Houston, TX 77005
*Corresponding to: Dr. Zeya Wang, Department of Statistics, Rice University, 6100 Main St. Duncan Hall 2124, Houston, TX 77005; Tel: +1(832)576-7107.
Background: Autism spectrum disorders (ASD) are classified as neurodevelopmental disorders. The aim of this study was to investigate the genetic risk of ASD by systematically reviewing the published literature and performing a meta-analysis.
Method: A comprehensive search of electronic databases was completed using Illumina BaseSpace Correlation Engine. Seven ASD case/control bio-sets from three different studies were selected, including 61 ASD cases and 83 controls. The top ASD risk genes from meta-analysis were further analyzed with an online open source ASD genetic database. Pathway enrichment analysis (PEA) and network connectivity analysis (NCA) were conducted to identify potential functional association between novel target genes and ASD.
Results: Two novel genes (YBX3 and HSPA1A) were identified through the meta-analysis as top target genes for ASD. These genes play roles within multiple ASD genetic pathways, demonstrating solid connection with known ASD target genes. Moreover, NCA results revealed strong functional association between these genes and ASD.
Conclusion: This study identified known as well as novel ASD target genes and their functional pathways that influence the ASD pathogenesis. Our results may add new insights for the understanding of the genetic mechanisms of ASD.
Autism Spectrum Disorder (ASD) is the name given to a group of related developmental disorders, which is characterized by language impairments, social deficits, and repetitive behaviors. The disease affects about 1 out of 68 children and about 6 out of 1,000 of overall population worldwide [1]. The most widely reported male and female ratio for autism prevalence is approximate 4 to1 [1].
The specific causes of ASD have yet to be found. Many risk factors have been identified in the research that may contribute to the ASD pathogenesis [2, 3]. Results of family and twin studies suggest that genetic factors play a role in the etiology of autism [2]. Additionally, studies found that ASD has an estimated heritability of around 50 % [4, 5]. The prevalence rate of autism in siblings of autistic children is about 15 to 30 times greater than that in the general population [6]. In addition, twin study showed that the concordance rates in monozygotic twins are much higher than that in dizygotic twins [7]. Recently, multiple genetic data from brain regions both at the gene expression and DNA methylation levels were employed for the continued efforts to identify ASD genetic determinants [8-10]. These studies built a solid background for ASD genetic research, which could be leveraged for the discovery and evaluation of novel risk genes.
However, it is hard to come to a consistent conclusion as results are spread over a large number of independent studies, which were often lack of power due to limited sample sizes and sample specificities in terms of phenotype characteristics. For an instance, the study to test transcriptional and epigenetic associations between brain regions and Autism conducted by Ginsberg et al., 2012 only employed data from 9 autism and 9 control subjects [8]. Therefore, a meta-analysis of multiple studies may provide a better assessment of the genetic risk factors of ASD with higher statistical power.
In this study, a meta-analysis was performed based on seven case/control bio-sets from three recent studies (2011 - 2012). The purpose of this study is to identify potential novel genes leveraging the power of meta-analysis. The top genes from the study were further analyzed integrating a curated ASD genetic database (ASD_GD_Feb012017). The ASD_GD_Feb012017 database was constructed using a large scale literature knowledge database, Pathway Studio (PS) ResNet database. In recent years, PS ResNet database has been widely used to study modeled relationships between proteins, genes, complexes, cells, tissues and diseases [http://pathwaystudio.gousinfo.com/Mendeley.html]. Our study identified novel ASD genes and suggested the effectiveness of integrating meta-analysis and PS ResNet database to identify and evaluate novel ASD risk genes.
This study tends to identify available gene expression case/control studies using data from brain issues for the meta-analysis. It is well accepted that expression data from brain regions are more relevant to metal health disorders like ASD than that from peripheral blood. A systematic search of available databases has been conducted using Illumine BaseSpace Correlation Engine (http://www.illumina.com). Figure 1 presents the diagram for the data selection. The initial search with target set as ‘Autism Spectrum Disorders’ identified 51 ASD studies. Further filter criteria includes: 1) The data organism is Homo sapiens; 2) The data type is RNA expression; 3) The samples of studies come from brain regions; and 4) The study is limited to ASD case vs. healthy control study (or include case/control bio-sets). In total, 7 bio-sets (ASD case/control comparisons) from 3 studies satisfy the selection criteria and were included in this systematic review and meta-analysis.
2.2 Genetic database ASD_GD_Feb012017The ASD_GD_Feb012017 is ASD targeted knowledge database online available at ‘Bioinformatics Database’ (http://database.gousinfo.com/). The current version ASD_GD_Feb012017 is composed of 523 ASD target genes (ASD_GD_ Feb012017 → Related Genes), 124 pathways (ASD_GD_Feb012017 → Related Pathways), and 80 related drug (ASD_GD_Feb012017 → Related Drug). Also included in the databases is the information of supporting references for each ASDGene and ASD-Drug relation, including the titles and the sentences where the relation has been identified (see ASD_GD_Feb012017 → Ref for Related Genes and ASD_GD_Feb012017 → Ref for Related Drugs, respectively). This information could be used to located detailed description of how a candidate gene/drug is related to ASD.
Using ASD_GD_Feb012017, network connectivity analysis was conducted to identify possible functional association between ASD and top target genes revealed by the meta-analysis. These analysis include identifying target genes related ASD pathways, genes and drugs. Here, we define two genes are functionally related if they play roles within same genetic pathway. Pathway enrichment analysis (PEA) was conducted using Pathway Studio (www. pathwaystudio.com) to identify genetic pathways potentially linked to ASD [11]. The gene-drug relation were identified using network building module of Pathway Studio.
2.3 Meta-score for Gene ResultsThis study uses gene meta-analysis score and specificity defined by BaseSpace Correlation Engine of Illumina (https://www.illumina.com/informatics/research/biological-data-interpretation/nextbio.html) to rank the genes from the meta-analysis and select the top significant genes. A gene's score is based on the statistical significance and consistency of the gene across the queried biosets. Correlation Engine assigns a numerical score of 100 to the most significant gene. All other scores are then normalized to the top-ranked gene (Correlated genes are ranked by specificity) and score 85 is considered as significant. A gene's specificity is the number of biosets in which the direction of a gene's regulation matches the selected filter.
After screening against the selection criteria, 7 ASD case/control comparison bio-sets from 3 independent studies were retrieved and assessed for eligibility. The first one contains one bio-set[9]. The second study contains 2 case/control bio-sets [8], including: 1) Cerebellum of autistic patients vs. normal controls; 2) Occipital lobe of autistic patients vs. normal controls. The third one contains 4 separate case/ control bisets [10], including: 1) Brain samples FFPE DASL assay - Autistic patients vs. healthy controls; 2) Brain samples FFPE IVT assay - Autistic patients vs. healthy controls; 3) Brain samples frozen DASL assay - Autistic patients vs. healthy controls; 4) Brain samples frozen IVT assay - Autistic patients vs. healthy controls. These three datasets were available at NCBI GEO (ID: GSE30573, GSE38322 and GSE28475, respectively), and the statistics of the included bio-sets are presented in Table 1.
As shown in Table 1, the case/controls for the three data sets were 19/17, 9/9, and 33/57, respectively. Further detailed information of these studies is available at GSE30573, GSE38322 and GSE28475.
The Meta-analysis results were curated as a signal file named as ASD_Meta and was deposited into the ‘Bioinformatics Database’ (http://database.gousinfo.com). Two genes, YBX3 and HSPA1A, passed the significant meta-analysis gene score (Score > 85; see Table 2). More detailed statistics were presented in ASD_Meta → Top 2 Genes. The full Metaanalysis results are presented in ASD_Meta → Full Gene List. A gene's score is defined by the metaanalysis Illumina BaseSpace Correlation Engine (http://www.illumina.com). The higher the score, the more importance of the gene for the case/control comparison.
YBX3 and HSPA1A were not included in in ASD_ GD_Feb012017, which indicates that these two genes could be novel significant genes for ASD. Therefore more attention should be paid to these genes with further analysis to be conducted. Analysis using the ASD_GD_Feb012017 showed that, these 2 genes were enriched within multiple ASD target pathways, and were connected to many other genes that were linked to ASD (see Table 2). Figure 2 (a) presents the functionally connections between these 2 genes and 158 ASD genes from ASD_GD_ Feb012017àRelated Genes. Figure 2 (b) presents the 9 ASD pathways (ASD_GD_Feb012017àRelated Pathways) including the 2 genes.
Functional network connectivity analysis (NCA) using Pathway Studio (www.pathwaystudio.com) showed that, the 2 novel genes from this meta-analysis (YBX3 and HSPA1A) present strong functional association with ASD. These genes influence the pathogenic development of ASD through multiple pathways, as shown in Fig. 3. Under each relation (arrow) in Fig. 3, there are support from one or more references (see ASD_Meta → YBX3, ASD_Meta → HSPA1A), which provide detailed description of each relation.
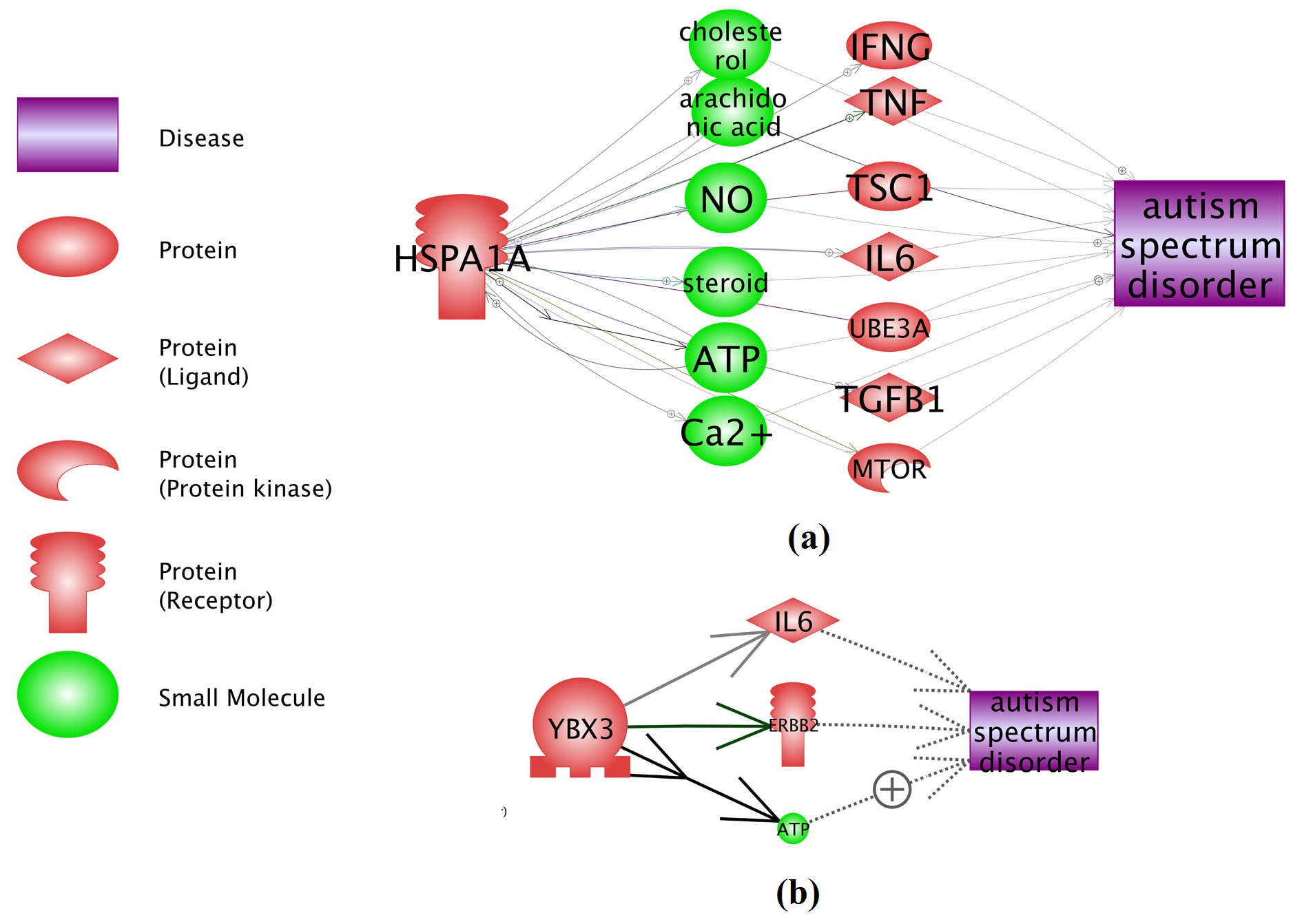
Fig 3. Network connectivity analysis between YBX3 and HSPA1A and ASD. (a) HSPA1A → ASD; (b) YBX3 → ASD. The networks were generated using ‘network building’ module of Pathway Studio. For the definition of the entity types and relation types in the figure please refer to http://pathwaystudio.gousinfo.com/ResNetDatabase.html
Meta-analysis is a statistical approach that combines the results of multiple scientific studies. Although many genetic studies have been conducted to discovery genetic risk factors for ASD, combine the results from these separated studies by using metaanalysis could lead to a higher statistical power and thereby more robust point estimate. In this study, meta-analysis was performed on 7 ASD case/ control bio-sets extracted from 3 recent studies. The target genes from meta-analysis were sorted by gene score, which is based on the statistical significance and consistency of the gene across the tested bio-sets. According to a recently updated database ASD_GD_Feb012017, meta-analysis results suggested two novel risk genes (i.g. YBX3 and HSPA1A) for ASD (Score > 85). These two genes were not previously implicated with ASD and yet among the top results from this meta-analysis. This suggested the necessity of further investigation of the potential functional relation between these two genes and ASD. In this study, multiple-level analysis has been conducted toward this purpose.
PEA results showed that, both YBX3 and HSPA1A are enriched within multiple ASD pathways (ASD_GD_Feb012017 → Related Pathways). Playing roles within these genetic pathways, these two genes are linked to dozens of other ASD genes (see Table 2 and Figure 2). These results support the possible association between these genes and ASD.
Additional network connectivity analysis (NCA) revealed multiple possible functional associations between ASD and the two novel genes (see Figure 3). It has been shown that YBX3 decreases IL-6 secretion [12], and IL-6 is a diagnostic marker and therapeutic intervention point for ASD [13, 14]. This suggests that YBX3 may play a role in the development of ASD through a YBX3 → IL-6 → ASD pathway. On the other hand, HSPA1A are involved in cholesterol transport [15]. Treatment with cholesterol supplementation in children with Smith- Lemli-Opitz syndrome has been reported to reduce the risk of ASD [16]. Thus, HSPA1A may influence the ASD pathogenesis by regulating cholesterol transportation. More potential regulation pathways could be identified from the ASD_Meta database (see ASD_Meta → YBX3, ASD_Meta → HSPA1A), which was curated from the results of this meta-analysis and has been deposited into the open source ‘Bioinformatics Database’ (http://database.gousinfo.com).
To sum up, this meta-analysis revealed two novel potential risk genes (YBX3 and HSPA1A) for ASD. Pathway enrichment analysis and functional network connectivity analysis support the meta-analysis results. This study also suggested possible functional pathways and mechanisms, thorough which these two genes may exert influence on ASD. Findings in this study may add new insights into the current field of ASD genetic study.
Authors claim no conflict of interests.
1.
2.
3.
4.
5.
6.
7.
8.
9.
10.
11.
12.
13.
14.
15.
16.
Liu D, Wang Z. Identification and Validation Novel Risk Genes for Autism Spectrum Disorder – A Meta-Analysis. J Psychiatry Brain Sci. 2017; 2(1): 2; https://doi.org/10.20900/jpbs.20170002

Copyright © 2020 Hapres Co., Ltd. Privacy Policy | Terms and Conditions